Research activity on systems radiation biology
The work carried out within this topic aims to study the effects of radiation on biological systems (e.g. cells, tissues, animals), integrating the different intra- and extra- cellular cascades and evaluating the perturbation induced to the systems as a function of dose delivered and time after irradiation.
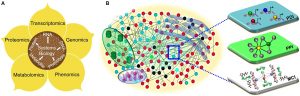 From: Front. Plant Sci., 30 June 2014
From: Front. Plant Sci., 30 June 2014
“Systems Biology” definition is not unique and rather multi-faceted, although can be probably described as an integrated, experimental and theoretical, approach which could build the basis for understanding and explaining the complexity of the biological systems investigated.
Why the Systems Biology approach is favourable?
The reason is straightforward: biological observations often suffer from the lack of quantitative approaches which could allow their computational analysis. If biology can be seen as an experimentally driven science, because investigated processes are not completely understood to allow for the implementation of mathematical models, systems biology is experimentally driven, computationally driven and knowledge driven [Szallasi et al.].
Systems Biology is grounded in the idea that the knowledge of a biological system cannot be extrapolated by the study of every single sub-module alone (reductionist approach) while only the study of the biological system as a whole can really fully explain the functions and behaviour (holistic approach).
Aims of Systems Biology approaches are to identify and mathematically model emergent properties in cells, organs, etc which cannot be measured or explained from the study of its single parts.
With the huge production of biological data coming from the new “omics” technologies (e.g. Next Generation Sequencing), the possibilities to understand biological systems as a whole are increasing day after day. Nowadays, all different molecular layers within the cells can be investigated in depth (genomics, transcriptomics, proteomics, etc) allowing a complete reconstruction of the metabolic and cellular networks more easily than before.
The research studies carried on within this topic mainly aims at unravelling the perturbation induced by the ionizing radiation insult on in vitro (cell cultures) or in vivo (animal) models. The datasets collected (either from our lab, from collaborations with other national/international groups or from publically available databases) are analyzed through the use of proper bioinformatic algorithms (mainly using the free software environment R).
Aims are either to find the molecules driving a well-known macroscopic effect (e.g. increased incidence of a cancer), exploring the altered biological pathways and functions (through the use of pathway analysis tools and software, e.g. Cytoscape) or understanding the non-linear response of all the molecular network (or a subset of it) in a temporal dynamic study (e.g. protein expression levels over time in cells acutely or chronically irradiated).
These studies are performed in close collaborations with other groups at the University of Pavia (e.g. Medicine department), Italian Universities (e.g. University of Verona) and research centres (ENEA-Casaccia, Rome) as well as European (e.g. Helmholtz Zentrum Munich, University of Stockholm, RIVM-Utrecht, University of Thessaloniki).
Systems Radiation Biology studies have been funded by European Projects such as DoReMi, EpiRadBio and OPERRA-Soprano.
For further reading:
- Z. Szallasi, J. Stelling, V. Periwal, “System Modelling in Cellular Biology: From concepts to nuts and bolts”, The MIT Press, 2010.
Links:
R – https://www.r-project.org/
Cytoscape – http://www.cytoscape.org/
ENEA Casaccia – http://www.enea.it/it/centro-ricerche-casaccia/
HMGU – https://www.helmholtz-muenchen.de/
RIVM – http://www.rivm.nl/en
SCK-CEN – https://www.sckcen.be/
Aristotle University of Thessaloniki – AUTH – https://www.auth.gr/en
Stockholm University – SU – http://www.su.se/english/